from IPython.display import YouTubeVideo
from datetime import timedelta
def insert_video(video_id, start=0, end=0):
if (start==0)and(end==0):
return YouTubeVideo(video_id,width=600, height=400)
#TO DO: use link "https://www.youtube.com/watch?t=19&v=WwoTzvOkEJQ" to get the id
start_seconds=int(timedelta(hours=start[0], minutes=start[1], seconds=start[2]).total_seconds())
end_seconds=int(timedelta(hours=end[0], minutes=end[1], seconds=end[2]).total_seconds())
return YouTubeVideo(video_id,width=600, height=400,start=start_seconds,end=end_seconds)
from IPython.display import HTML
HTML('''<script>
code_show=true;
function code_toggle() {
if (code_show){
$('div.input').hide();
} else {
$('div.input').show();
}
code_show = !code_show
}
$( document ).ready(code_toggle);
</script>
<form action="javascript:code_toggle()"><input type="submit" value="Click here to toggle on/off the raw code."></form>''')
Block 1
Block 1¶
Lecture 1. Gene Expression¶
Part 1. Introduction¶
DNA is information storage
- reading the information (gene expression)
- passing the information on
- in dividing cells: from cell to cell
- in reproduction: from parents to offspring
Chemical bounds
- covalent bonds:
- share electrons between atoms
- strong bonds
- bonds within most organic compounds
- hydrogen bonds:
- two atoms share a hydrogen atom (H)
- usually weaker then covalent bonds
- donor: N or O atom with covalently linked H
- acceptor: O or N
Nucleotides
- carrier of chemical energy (ATP)
- cofactors of enzymes
- specific signaling molecules in cells
- building blocks of nucleic acids ?
DNA
- Complementary Base Pairing (A=T, G=C by hydrogen bounds )
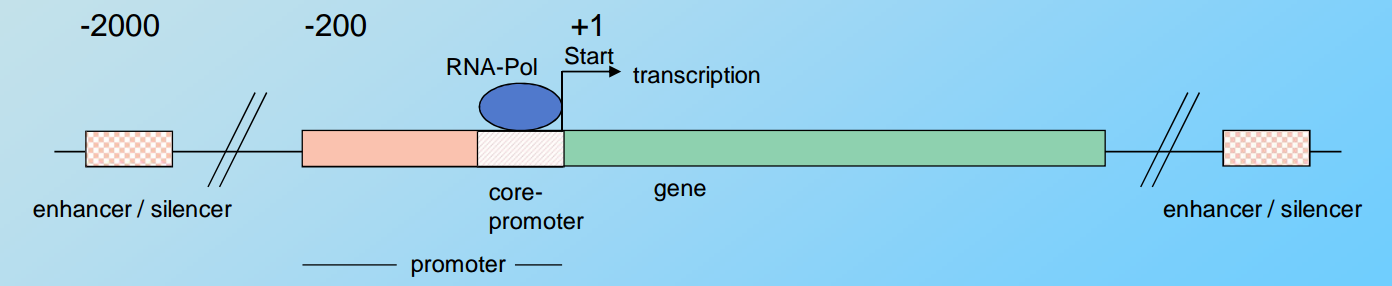
Gene
DNA sequence that is transcribed as a single unit and encodes one protein (or a closely related set of proteins, see: alternative splicing)
- eucaryotic genes contain exons and introns
- exons are the protein coding regions
- introns are removed from the mRNA by splicing
Chromosomes
Eucaryotic DNA is Packaged into a Set of Chromosomes
- Chromatin: the complex of DNA and protein
Replication
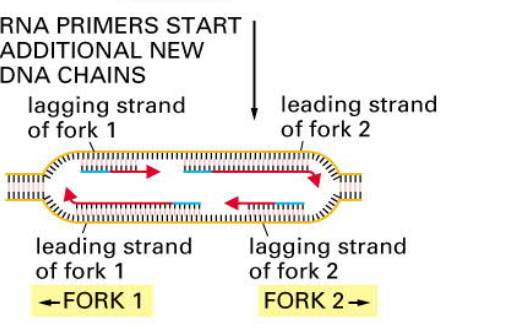
How to copy DNA?
- leading strand: is synthesized continously (3 -> 5 in template)
- lagging strand: is synthesized discontinously by a „backstiching“ mechanism ( 5->3 in template, but reconstraction is in 3->5 direction )
- Okazaki Fragments: short DNA fragments synthesized at the lagging strand, are later joined together by the enzyme DNA ligase
- You need initiation protein to open double helix
- replication starts with RNA primers
- replication of chromosomes bidirectionally in replication bubble
Part 2. Transcription¶
RNA
Types of RNA
- mRNA: messenger RNA is translated to produce a protein
- tRNA: transfer RNA adaptors in protein synthesis (translation)
- rRNA: ribosomal RNA part of the ribosomes
- miRNA: micro RNA regulatory functions
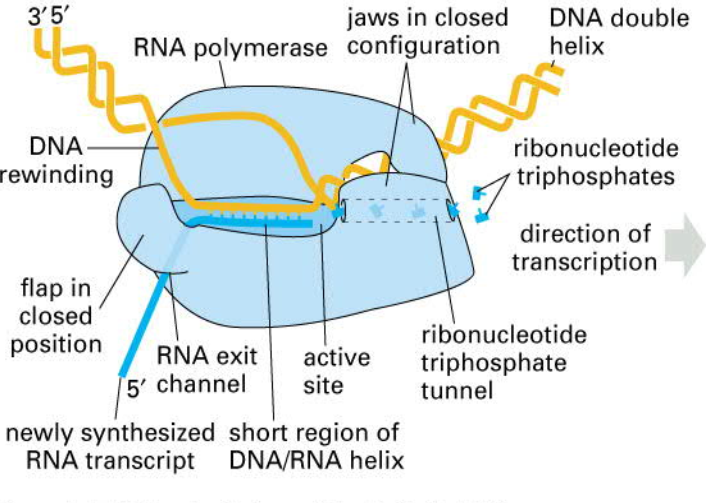
RNA POL (binds to the promoter)
Types of RNA polymerases
- RNA Pol I makes the large ribosomal RNAs
- RNA Pol II transcribes genes into mRNA
- RNA Pol III makes a variety of small RNAs (5S rRNA, tRNA)
- new mRNA is generated in 5‘ to 3‘ direction
- stops when "see" secondary structure in RNA that destabilizes the hold of the RNAPol on the RNA
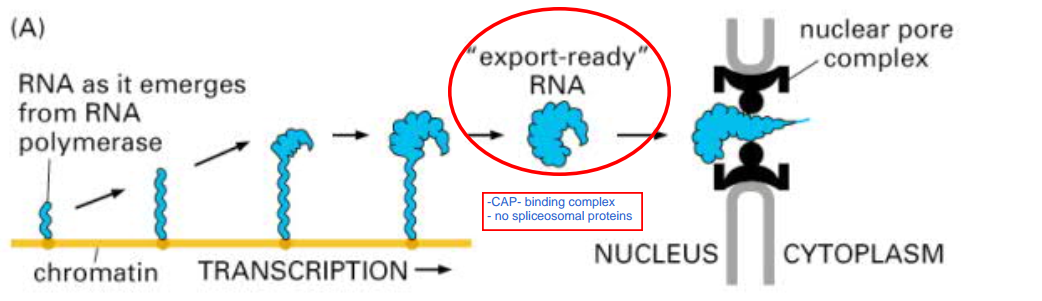
RNA Splicing
- alternative splicing can lead to protein variants made from the same gene:
- different exons from one gene can be joined together
- the presence of numerous introns in DNA allows genetic recombination to combine the exons of different genes
- important process for evolution of new proteins from part of existing ones
Exporting
Part 3. Translation¶
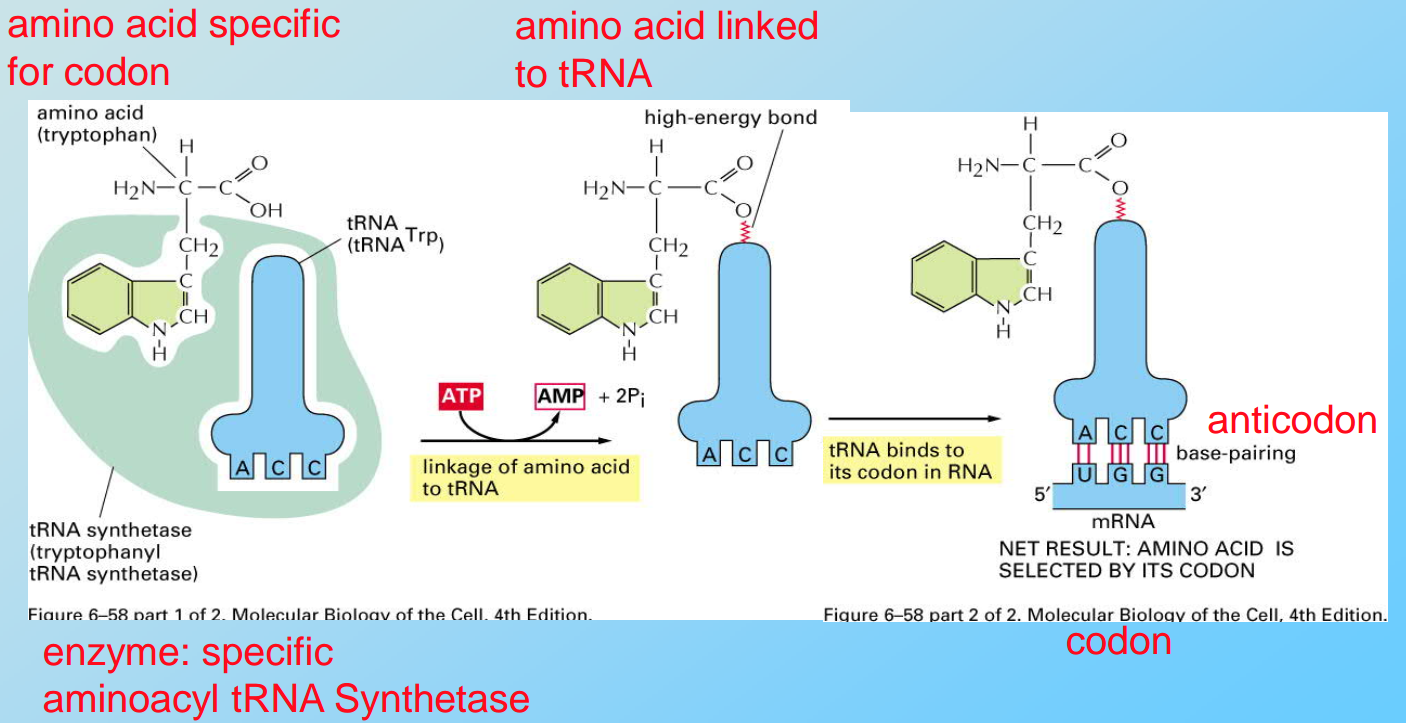
The Genetic Code
Three nucleotides (a triplet) code for one amino acid
- different triplets can code for the same amino acid e.g. GCA, GCC, GCG, GCU code for alanine
- AUG codes for methionine at the translation start and within the protein
- UAA, UAG and UGA are stop codons
- frameshift mutation = 1 or 2 nucleotides deleted or inserted (leads to a totally different protein)
- change in one gene can lead to AD
Mechanism
Initiation:
- initiationsignal is the first AUG following the 5’-Cap of the mRNA
- initiator tRNA (tRNAi) is coupled to methionine
- “scanning”: small ribosomal subunit with tRNAi and eucaryotic initiation factors (eIFs) binds to 5’-Cap and scans along the mRNA for the start codon, then the large ribosomal subunit is recruited
Elongation:
- coupling of amino acids to the growing chain is mediated by elongation factors
- each coupling reaction is associated with GTP hydrolysis to GDP
Termination:
- a release factor binds to the Stop-codon and terminates translation
- the protein is released from the ribosome
Parallelism
- mRNA is translated by many Ribosomes in Parallel
Part 4. Regulation¶
- transcript gene or not
- how many mRNA to produce from one gene
- ofter low rate without additional factors (gene regulatory proteins)
- activators and repressors influence transcription rate
cis-regulatory elements:
- sequences in the DNA that control initiation and efficiency of transcription
- serve as high affinity binding sites for regulatory proteins called transcription factors
Lecture 2. Biological Membranes¶
Part 1. Structure¶
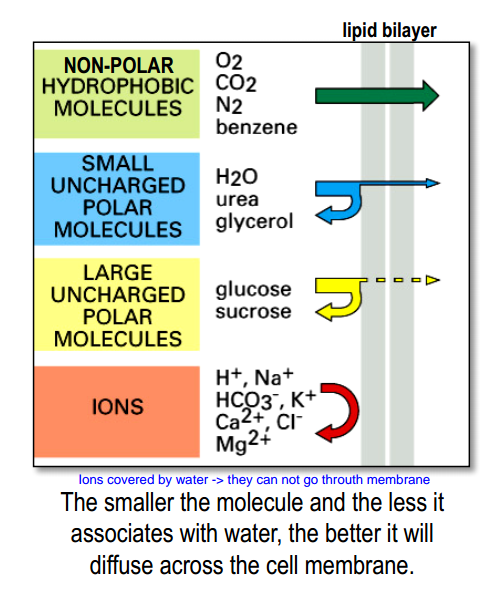
Functions of membrane
- plasma membrane defines the inside and the outside of a cell
- internal membranes form the boundaries of organelles
- prevent molecules generated inside the cell from leaking out
- prevent unwanted molecules from diffusing in
- contain transport systems, serve as transport system (vesicles)
- accomplish energy and compound storage
- perform information transduction
- mediate cell-cell contact
Cellular membranes are asymmetric
Loss of asymmetry functions as apoptotic signal
insert_video("h7x6Ojc_6iI")
Part 2. Membrane proteins¶
- helix transmembrane domain (mostly non-polar)
- membrane anchors (hydrophobic groups that are covalently attached to proteins)
Lipid Rafts
Summary
- Membranes control the energy, information and compound flow into and out of the cell and its compartments
- Membranes are asymmetric „2 dimensional liquids“
- Membranes contain specialized compartments lipid rafts
- Anchor points for cytoskeleton
- Preformed organized areas for signal transduction and cell cell contacts
- Integral and peripheral membrane proteins perform most of the tasks and help membrane organization
- Proteins may span the membrane to allow small charged molecules to cross the membrane
Part 3. Traffic¶
Protein sorting
Properties
- Proteins are synthesized in the cytoplasm and need to be transported to different cellular compartments
- Proteins contain all the information for proper targeting in their AA sequence in form of signal sequences
- Signal sequences are recognized by other receptor proteins that mediate the interaction with the translocation maschinery
- Protein translocation is energy consuming
insert_video("owdIv8M5wjA", start=(0,9,18),end=(0,14,53))
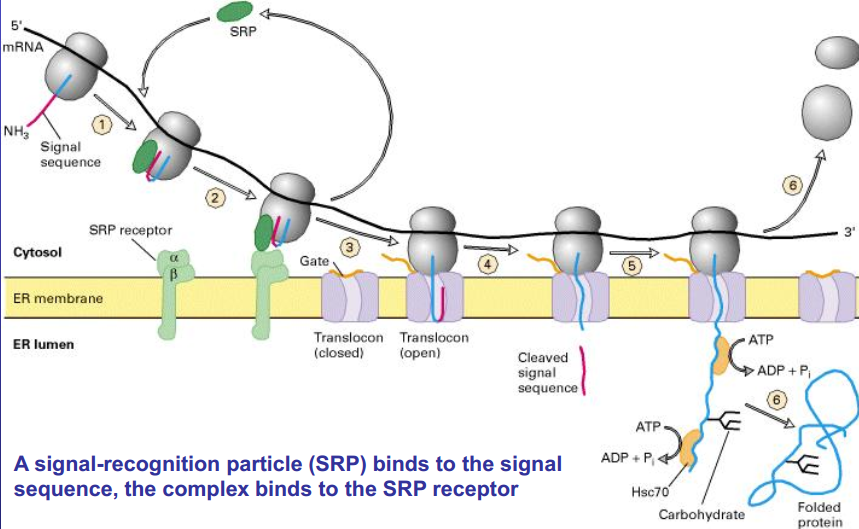
Synthesis of secretory proteins
Signal sequence of ER + SRP -> ribosome goes to rough ER -> SRP (and ribosome) binds to SRP receptor -> SRP is free -> ribosome moves to translocon and opened it -> signal sequence is removed -> production of protein into ER (hsc 70 prevent folding) -> after the whole protein is produced, it is folded.
Protein Translocation through the nuclear pore
insert_video("owdIv8M5wjA", start=(0, 4, 10),end=(0,9,18))
Summary
- The nuclear pore complex acts as a gated channel through which macromolecules are selectively transported in and out of the nucleus.
- Transport through the nuclear pore is active and directed.
- Proteins actively imported through nuclear pore complexes contain a nuclear-localization signal (NLS). Shuttling proteins contain both a nuclear-export signal (NES) and NLS.
- The NLS never gets cleaved off.
Lecture 3. Proteins¶
Part 1. Folding¶
Forces that establish a 3D-structure:
- ionic bond
- hydrogen bond
- wan-der-Waals attraction between atoms
So we try to minimize the hydrophobic surface
Structures
- Primary structure: amino acid sequence
- Secondray structure: highly regular local sub-structures (α-helix; β-sheet)
- Tertiary structure: 3-D structure of a single, protein molecule
- Quartery structure: 3-D structure of a complex of 3-D folded proteins together with auxiliary factors
To prevent folding until the end of transduction (hsp70), or change folding (hsp60)
incorrect folding can force other proteins fold incorrect (protein aggregates) this can cause diseases (AD, PD so on)
Part 2. Posttranslational Modifications¶
- Phosphorylation of Proteins at Serine, Threonine or Tyrosine Residues
- Kinases add phospho- group and phosphatases delete phospho- group
- can activate and inactivate protein
Posttranslational Modifications in Rough ER
Before reach destination:
- Formation of disulfide bonds
- Intramolecular and intermolecular disulfide bonds (Cys–S–S–Cys) help stabilize the structure of many proteins
- Disulfide bond formation can proceed spontaneously only when sufficient oxidant is present, in eukaryotic cells, disulfide bonds are formed in the lumen of the rough ER but not in the cytosol.
Proper folding (proteins that accelerate refolding)
- Protein disulfide isomerase (PDI)
- The chaperone Hsc70
Assembly into multimeric proteins
- Addition and processing of carbohydrates (glycosylation)
- Assist in folding
- Stabilization of glycoproteins
- Cell-cell adhesion
- Targeting signal (Mannose 6-phosphate is the marker that directs many hydrolytic enzymes from the Golgi complex to lysosomes )
- Specific proteolytic cleavages
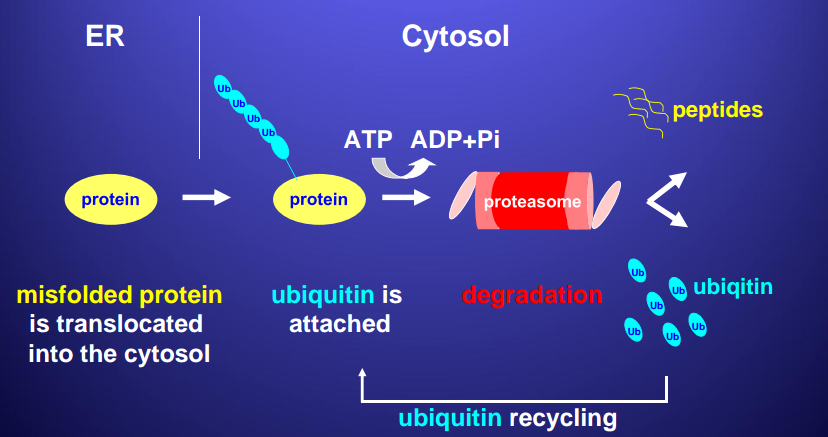
Part 3. Protein Degradation¶
The cell degrades:
- misfolded or denatured proteins
- normal proteins whose concentration must be decreased
- foreign proteins taken up by the cell
Life Span of Proteins
- sort (less than 3 minutes) e.g. regulatory proteins
- long (age of an organism for some structural proteins) e.g. proteins in the lens of the eye
- depends on amino acids in N-terminal
Lysosomes
- the acid hydrolases are hydrolytic enzymes that are active under acidic conditions
- the lumen is maintained at an acidic pH by an H+ ATPase in the membrane that pumps H+ into the lysosome
Proteasome
During unfolded protein stress (UPS) cell produces hsc70, enzymes to help proteasome, and proteins that move proteasome to ER
Block 2¶
Lecture 1. Neurotransmitter receptors¶
There are three major classes of cell-surface receptors:
- Ion-channel-linked receptors
- G-protein-linked receptors
- Enzyme-linked receptors (signal molecule activates enzyme)
Part 1. Ionotropic receptors¶
- Ionotropic (NT opens an ion channel) - fast, short-lasting synaptic transmission
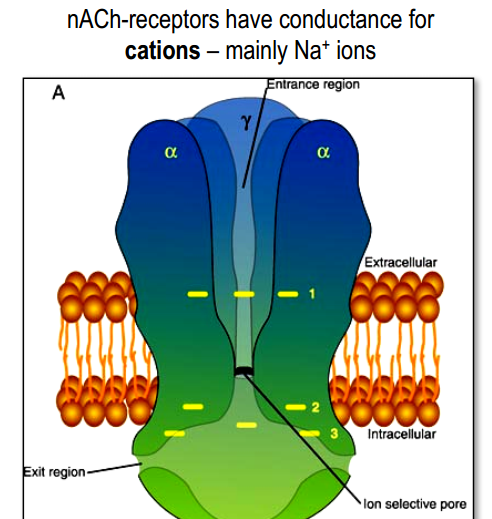
Example: ionotropic nACh-receptor
>
- 5 protein sub-units, each from 4 regions.
- M2 regions create pore.
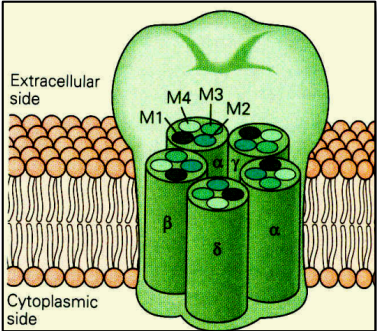 2 molecules of $ACh$ in $\alpha_{\gamma}$ and $\alpha_{\delta}$ sites
2 molecules of $ACh$ in $\alpha_{\gamma}$ and $\alpha_{\delta}$ sites
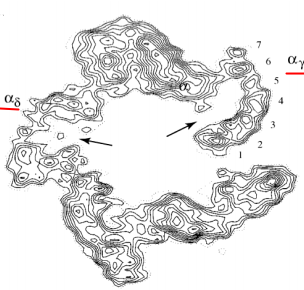
- 3 rings of negatively charged amino acids (Glu-, Asp-)
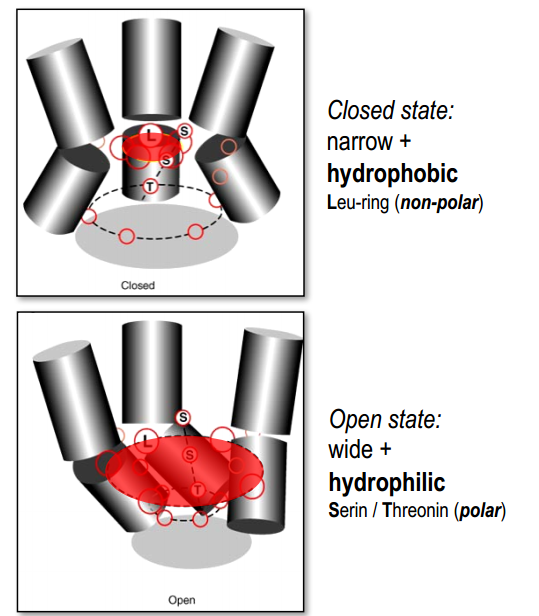
Binding of $ACh$ $\rightarrow$ rotation (conformational change) $\rightarrow$ opened state
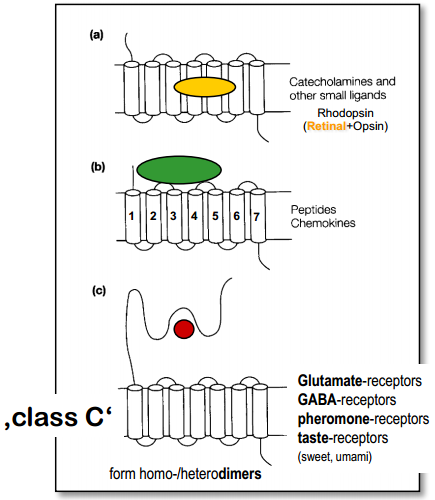
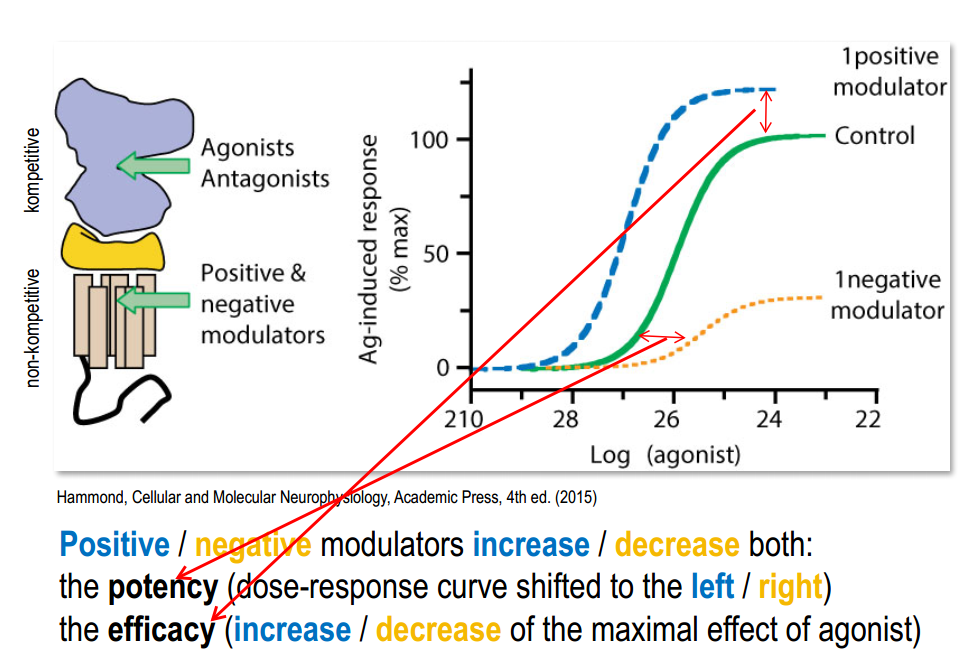
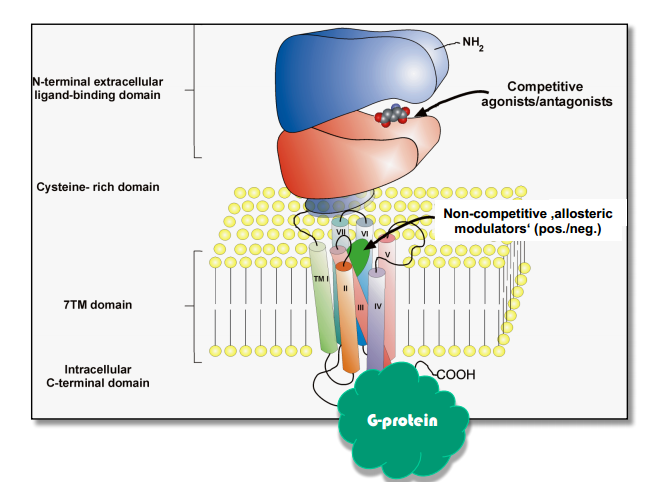
Part 2. Metabotropic receptors¶
- Metabotropic NT receptor (NT triggers intracellular processes) - slow, long-lasting
Classification (by binding)
- Binding site inside the membrane [Examples?]
- Binding site on the surface of membrane [Examples?]
- Binding site inside long N-terminal (venus flytrap module - VFTM)
Process:
Ligand binding $\rightarrow$ Conformational change $\rightarrow$ Binding & activation of G-protein
Activation of G-protein:
Activated $\alpha$ (with GTP) and $\beta \gamma$-subunits unbind from receptor $\rightarrow$ $\alpha$ (with GTP) binding to target protein and activate it $\rightarrow$ $\alpha$-subunit unbinds from target protein via energy from hydrolysis of GTP (GTP $\rightarrow$ GDP) $\rightarrow$ binding of $\alpha$ and $\beta \gamma$-subunits $\rightarrow$ inactivated G-protein.
Amplification:
- G-protein $\rightarrow$ $10^{10}$ Glycogen molecules
- One Rodopsin molecule $\rightarrow$ many $250 Na^+$ channels closed (rod cell hyperpolorized by $1 mV$)
Activation of different processes(coordinated modulation):
- metabolic enzyme $\rightarrow$ altered metabolism
- gene regulatory protein $\rightarrow$ altered gene expression
- cytoskeletal protein $\rightarrow$ altered cell shape and movement
Can be interaction between different processes triggered by second messengers (crosstalk)
Part 3. Calcium as second messenger¶
Functions
In neurons
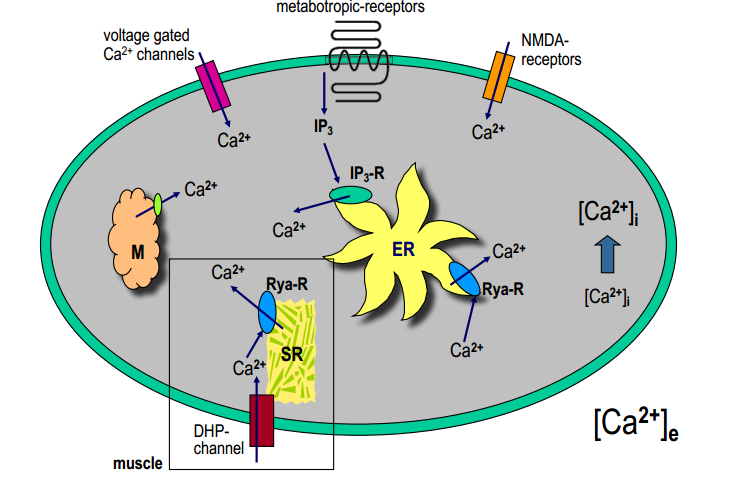
- neurotransmitter release at the chemical synapse
- gating of ion channels ( $Cl^-_{Ca}, K^+_{Ca}, Ca^{2+}_{Ca}$)
- gating of Ryanodin- and IP3-receptors ($Ca^{2+}$-induced $Ca^{2+}$-release)
- activation of various enzymes: $Ca^{2+}$-calmodulin, $Ca^{2+}$-protein kinases
- activation of transcription factors – gene expression
- involvement in ‘synaptic plasticity’ – LTP
In muscle cells
- electromechanical coupling – $Ca^{2+}$-binding to troponin C (de-blockade of a-m-binding) ???
- heart muscle cells: delayed $Ca^{2+}$-influx – prolongation of AP‘s
Related problems
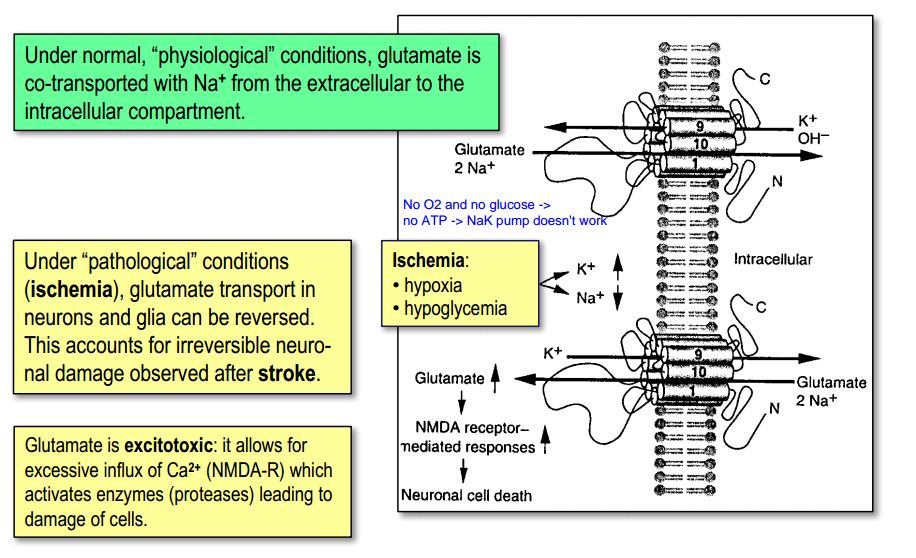
- increased $[Ca^{2+}]_{int}$ is neurotoxic ! (Glu NMDA-rec.)
- dysregulation of $Ca^{2+}$ can lead to various pathologies
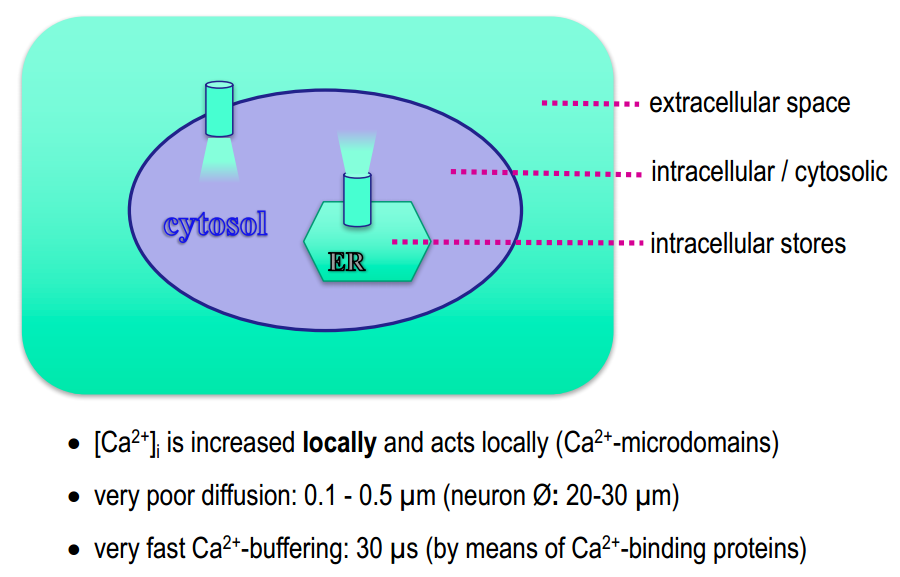
$Ca^{2+}$-compartments
$[Ca^{2+}]_{int}$ is very small and can change in 1000 times. So it have to be regulated precisely in space and time.
Getting out
$Ca^{2+}$-binding proteins : parvalbumine, calbindine (cytosol) calsequestrine (SER)
Mechanisms of $Ca^{2+}$-buffering
- $Na^+-Ca^{2+}$-transporter in plasma membrane: high capacity - low affinity (to quickly remove $Ca^{2+}$ when the concentration is big)
$H^+-Ca^{2+}$- in mitochondria transporter: high capacity - low affinity
$Ca^{2+}$-ATPase pump: low capacity - high affinity (works even when there is small $[Ca^{2+}]$)
- PMCA: plasma membrane $Ca^{2+}$-pump
- SERCA: smooth endoplasmic reticulum $Ca^{2+}$-pump
- MMCA: mitochondrial membrane $Ca^{2+}$-pump
Lecture 2. GABA-receptors¶
Part 1. Ionotropic GABA-receptors (GABA$_A$ and GABA$_C$)¶
Properties
- GABA$_A$-R and GABA$_C$-R are anion channels
- they have conductance for $Cl^-$ (influx)
- M2 forms the pore – „selectivity filter“
- M2 has rings of positive charge (Lys+, Arg+)
- the $\beta$-subunit binds GABA ($\alpha$-subunit help a little)
- needed 2 GABA molecules
- GABA$_A$-R bind additional substances(they modulate the channel properties)
Modulation of GABA$_A$-receptors (GABA$_C$ doesn't have such properties )
benzodiazepine binding (only with $\gamma$-subunit and only $\alpha_1$ has high affinity )
Experiment: Potentiation of GABA$_A$-receptor currents
- transfection of Xenopus oocytes with cDNAs of $\alpha$-, $\beta$-, $\gamma$-subunits
- recordings with whole-cell patch-clamp technique
- application of GABA alone
- co-application of GABA + benzodiazepines (diazepam - DZP, FNZM)
- co-application von GABA + barbiturates (pentobarbital - PB)
Result: benzodiazepines and barbiturates induce more inhibition The same result we can obtain by stimulation of GABA-ergic interneurons (pre-synapt.) in the hippocampus brain slice preparation
So we see potentiation of IPSP.
Part 2. Metabotrophic GABA-receptors (GABA$_B$ )¶
Properties
- 2 types: GABA$_B$1a-d and GABA$_B$2a-c (only 35% sequence homology)
- B1 / B2 are “co-distributed” in the brain (are even expressed in the same cells)
- expressed in all brain areas
- GABA$_A$-R vs. GABA$_B$-R: 50% / 50%
- in brainstem, thalamus, cerebellum, spinal cord: 90% GABA$_B$-R
- located pre-, post- und extra-synaptically
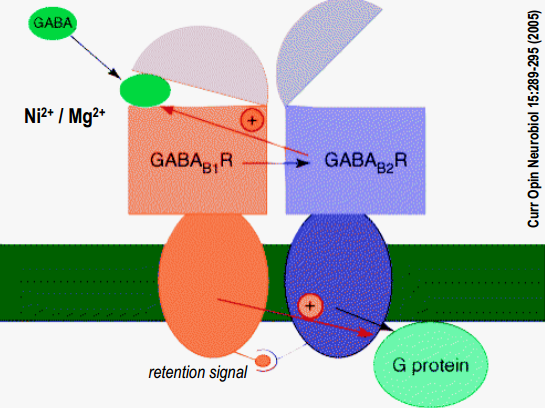
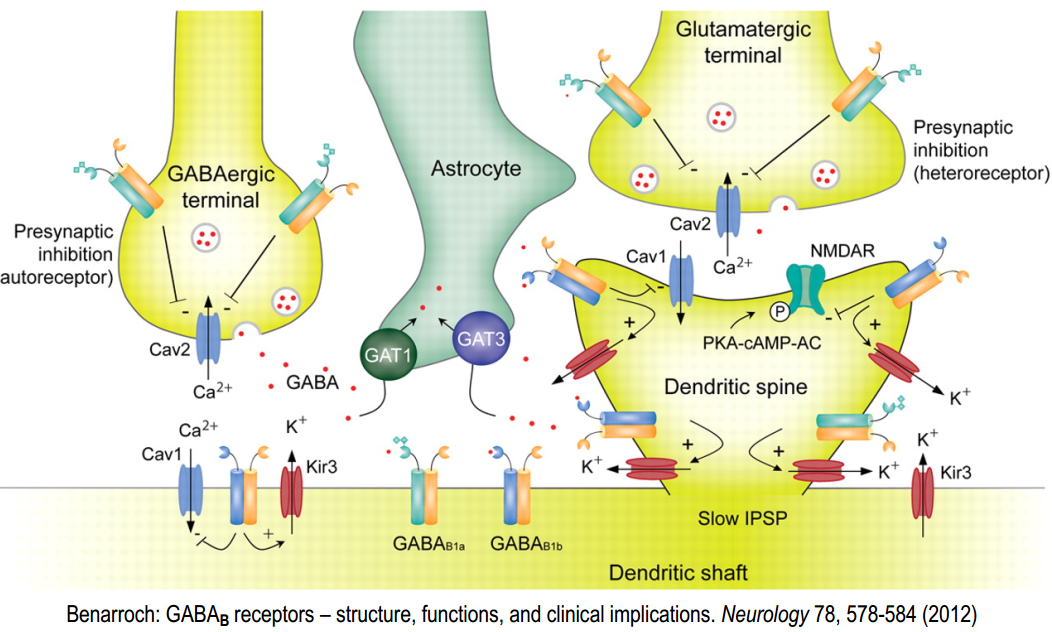
GABA$_B$-R form heterodimers
- B1 and B2 work together and have different functions (B1 ligand binding, B2 G-protein binding)
GABA$_B$-receptor: effector systems
- GABA$_B$-receptor binds i/o-G proteins, that have such effect:
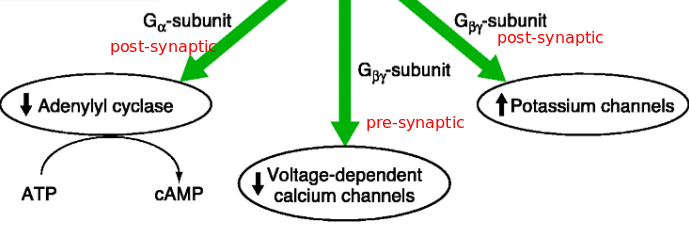
Post-synaptical ($\alpha$-subunit):
Activation of GABA$_B$-R post-synaptically activ. Gi/Go $\alpha$-SU inhibits AC $\rightarrow$ reduction of cAMP-level $\rightarrow$reduction of cAMP-dependent processes $\rightarrow$ reduction of protein kinase A-level (PKA) $\rightarrow$ no PKA – no phosphorylation
Post-synaptical ($\beta \gamma$-subunit):
activ. GABA$_B$-R post-synaptically $\rightarrow$ activ. Gi/Go (separation of $\alpha$-SU + $\beta \gamma$-SU) $\rightarrow \beta \gamma$-SU binds to $K^+$-channel (GirK) $\rightarrow$ gating: $K^+$ outflow $\rightarrow$ hyperpolarisation
Pre-synaptic effector systems (can be done by GABA released by the same synapse (auto receptor), or by other neuron that inhibit synapse (hetero receptor) )
activ. GABA$_B$-R post-synaptically $\rightarrow$ activ. Gi/Go (separation of $\alpha$-SU + $\beta \gamma$-SU) $\rightarrow$ $\beta \gamma$-SU binds to voltage gated $Ca^{2+}$ channels $\rightarrow$ modifies voltage dependence $\rightarrow$ no gating – no/reduced $Ca^{2+}$-influx in pre-synapse $\rightarrow$ no/reduced transmitter-release
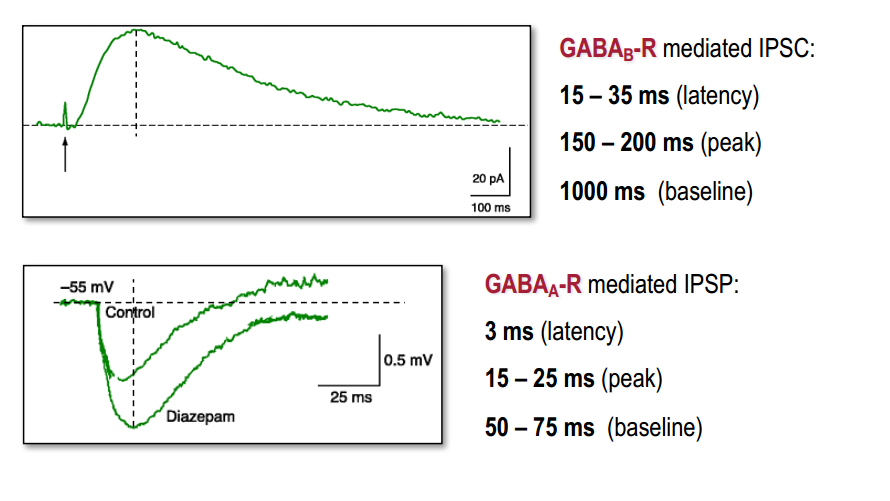
Time course
Summury
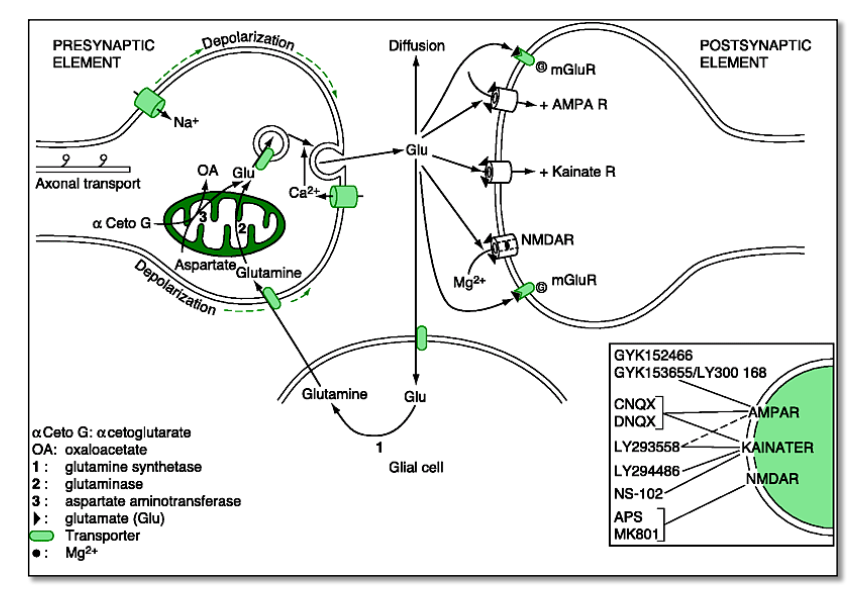
Lecture 3. Glutamate receptors¶
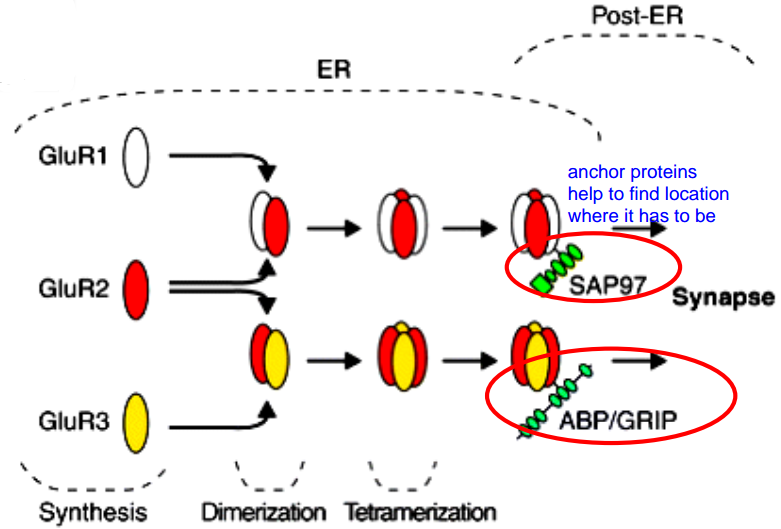
Part 1. Ionotropic Glu-receptors (AMPA-Kainat-R, NMDA-R)¶
Properties
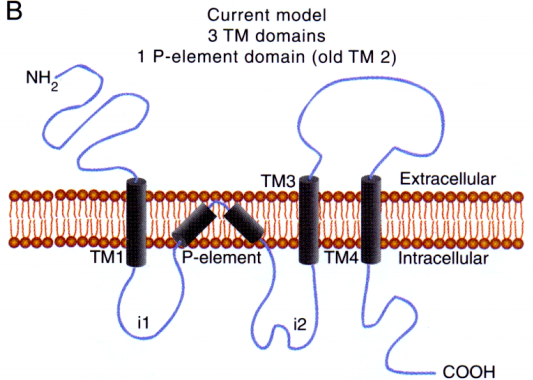
- iGlu-Rs have been ‘pharmacologically’ differentiated based on their specific synthetic agonists and antagonists
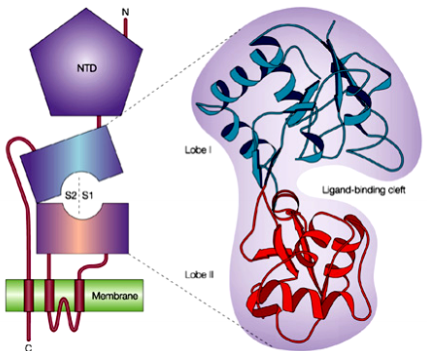
- iGlu-Rs have different molecular structure (3 transmembrane domains
- 1 pore-element)
- They first assemble to form a dimer, then two dimers form a tetramer
Ligand binding
- Ligand binding occurs in the cleft formed by lobe I + lobe II. The two lobes are collectively formed by the extracellular loops.
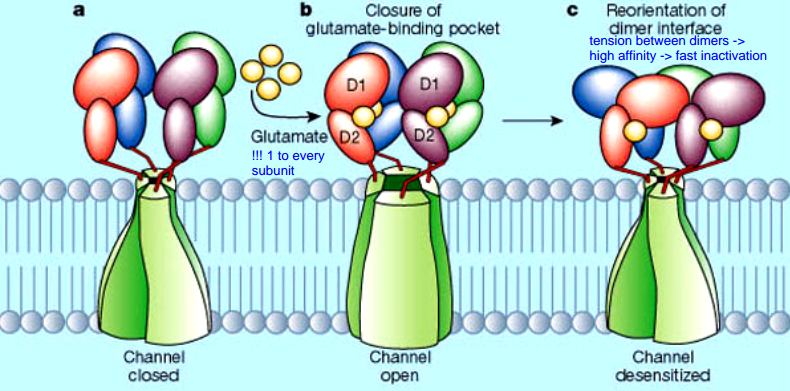
- 4 Glu to one iGlu-R
- Glu binding and subsequent conformational change causes tension between dimers
- affinity of the dimer-interfaces determines the inactivation (desensitization):
- high affinity $\rightarrow$ fast inactivation $\rightarrow$ small current (iGlu-R flop)
- weak affinity $\rightarrow$ slow inactivation $\rightarrow$ large current (iGlu-R flip)
Part 2. NMDA¶
We need two receptors to activate NMDA
- Glycine (or ‘D-serine’) is bound by NR1 subunits (S1-S2-half domains)
- Glutamate is bound by NR2 subunits (S1-S2-half domains)
Other properties:
- NMDA-Rs are cation channels with high $Ca^{2+}$-conductance
- play a major role in „synaptic plasticity“
- play a role in „glutamate-toxicity“ – [$Ca^{2+}$] (when glutomate stay in extracellular space $\rightarrow$ too many $Ca^{2+}$)
- NMDA-Rs are „ligand gated“ and „voltage gated“
- gating requires pre-depolarization – removes ‘Mg2+-block’(accomplished by fast AMPA-Rs) )
- NMDA-Rs open and close ‘slowly’ – late EPSP phase
- NMDA-Rs have several modulatory binding sites (hallucinogenic drugs(PCP), MK801, Ketamine, polyamines, pH (H$^+$))
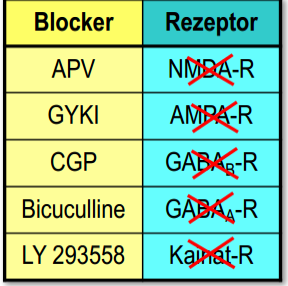
Experiment:
To check that NMDA-Rs close slowly, we can block it and look for difference
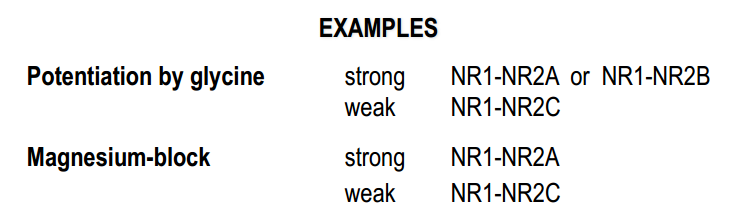
Useful table for such experiments
NMDA-R subunits and channel properties
- functional channels are hetero-tetramers
- they consist of NR1 -+ NR2-subunits (type of NR2 is important for functions)
- NR1-NR3B-heteromers form cation channels which are gated by glycine alone. (excitatory glycine receptor, no $Ca^{2+}$-conductance, no Mg2+-block, expressed in the spinal cord )
Summary
Part 3. Metabotropic Glu-R¶
mGlu-Rs are the most important neuromodulators in the brain¶
mGlu-Rs are most abundant in the brain¶
Classification(3 groups confirmed):
- sequence homology(Group I: R1, R5; Group II: R2,R3, Group III: R4, R6, R7, R8)
- pharmacologically
- by G-prot. / sec.mess. couplings(Group I: $G_q$; Group II:$G_i$ Group III: $G_i$ (the same as in GABA$_B$))
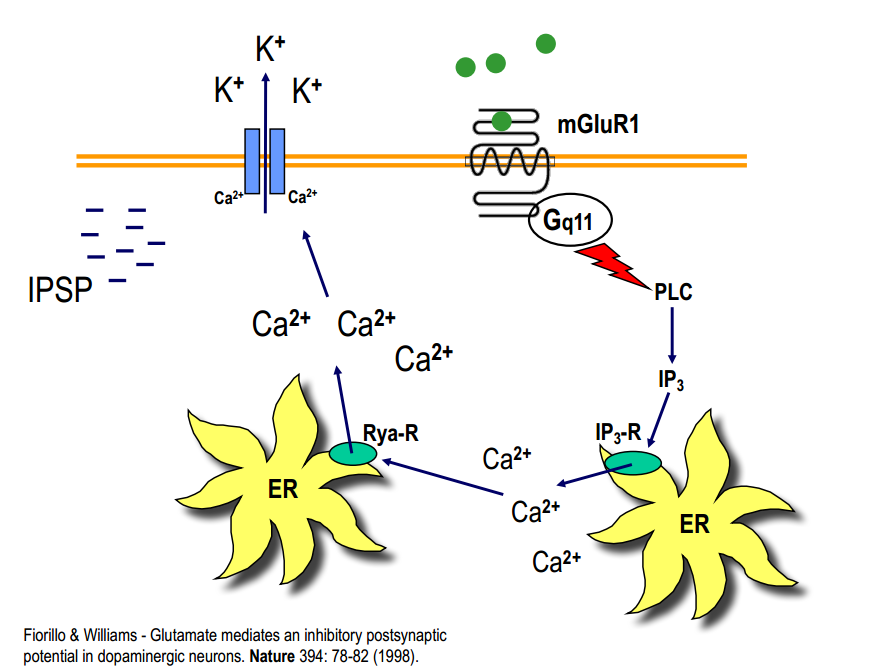
Modulation
Group I and $G_q$-protein
- activation of PLC $\rightarrow$ IP3 $\rightarrow$ $Ca^{2+}$ -release from intracellular stores
- mGlu-R1: single $Ca^{2+}$-pulses $\rightarrow$ activation of cation-channels or activation of $Ca^{2+}$-depend proteins
- mGlu-R5: $Ca^{2+}$-oscillations (2/min) $\rightarrow$ activ. transcription factor $\rightarrow$ gene expression
- activation of PLC $\rightarrow$ DAG $\rightarrow$ activation of PKC $\rightarrow$ phosphorylation of target proteins $\rightarrow$ activ. MAP kinase $\rightarrow$ transcription factor $\rightarrow$ gene expression
mGlu can induce IPSP
Lecture 4. Neurotransmitter Transporters¶
Part 1. Introduction¶
Cell membrane is a barrier for many molecules
- Barrier allows for intra- vs. extracellular concentration differences $\rightarrow$ resting potential
- Barrier allows for precisely controlling what goes in-and-out of a cell.
- Up to 30% of all genes code for transport proteins.
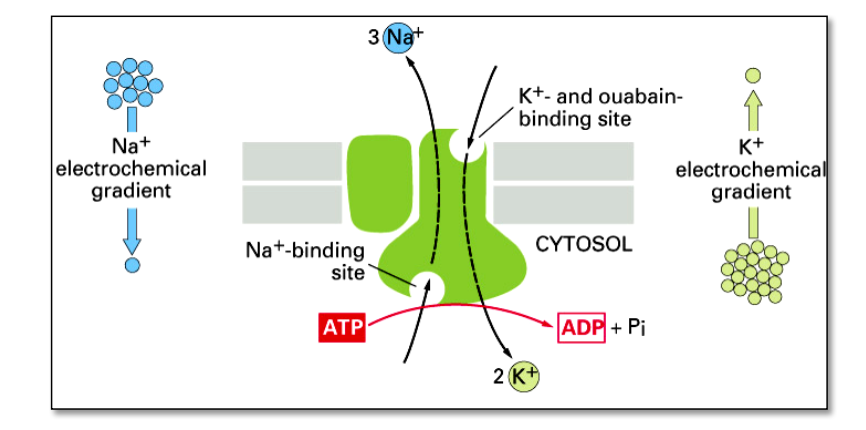
- Neurons devote the majority of their metabolic energy consumption to membrane transport mechanisms ($Na^+-K^+$ pumps burn up to 2/3 of the energy of neurons)
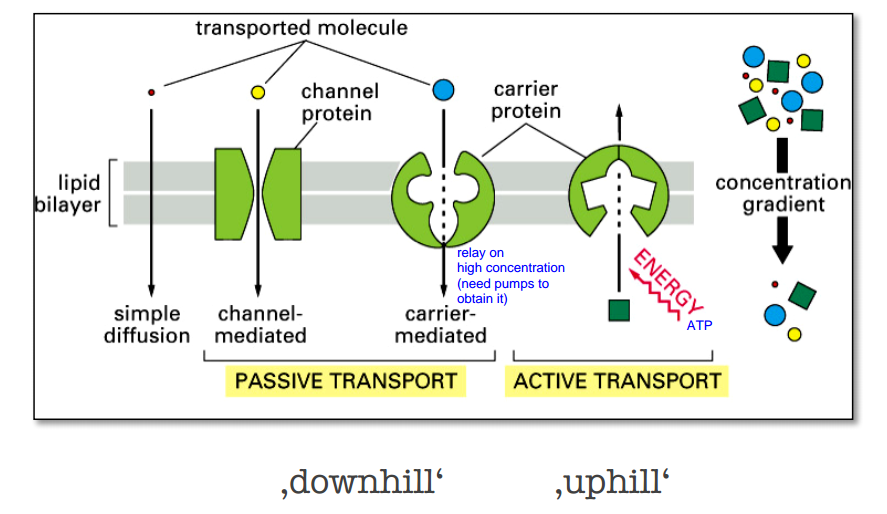
Passive transport vs Active transport
Active transport
the $Na^+-K^+$ pump actively pumps $Na^+$-ions out of the cell and $K^+$-ions into the cell – against their concentration gradients.
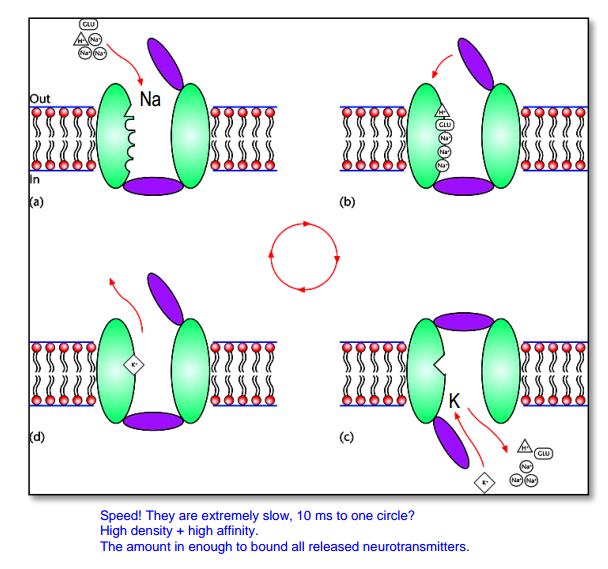
Part 2. Neurotransmitter (re)-uptake¶
Every part of synapse has NT transporter. Vesicles also have( transport against their gradient).
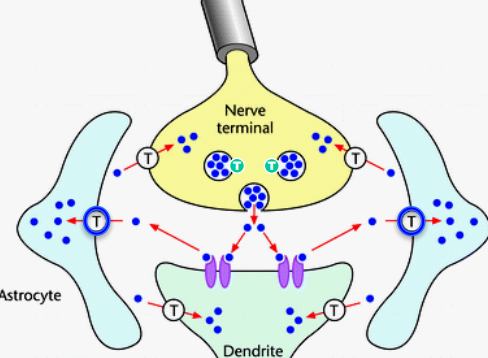
- Transporters are slow (takes several 10 ms for 1 transport cycle)
- BUT to compensate this:
- NT transporters are: expressed abundantly / in high density
- transporters bind NT with high affinity
- The number of „high affinity binding sites“ (= NT-receptors + NT-transporters) is such that NT-molecules from 1 release event is completely bound
- The NT-transporters are ‚done‘ with a cycle and ready again when NT-molecules are unbound from the NT-receptors (so all molecules turn back in intracellular space of post-, pre-synaptic site or astrocyte )
- So this makes NT short in time. So EPSP is near in time with AP and we have communication
SLC(solute carrier)
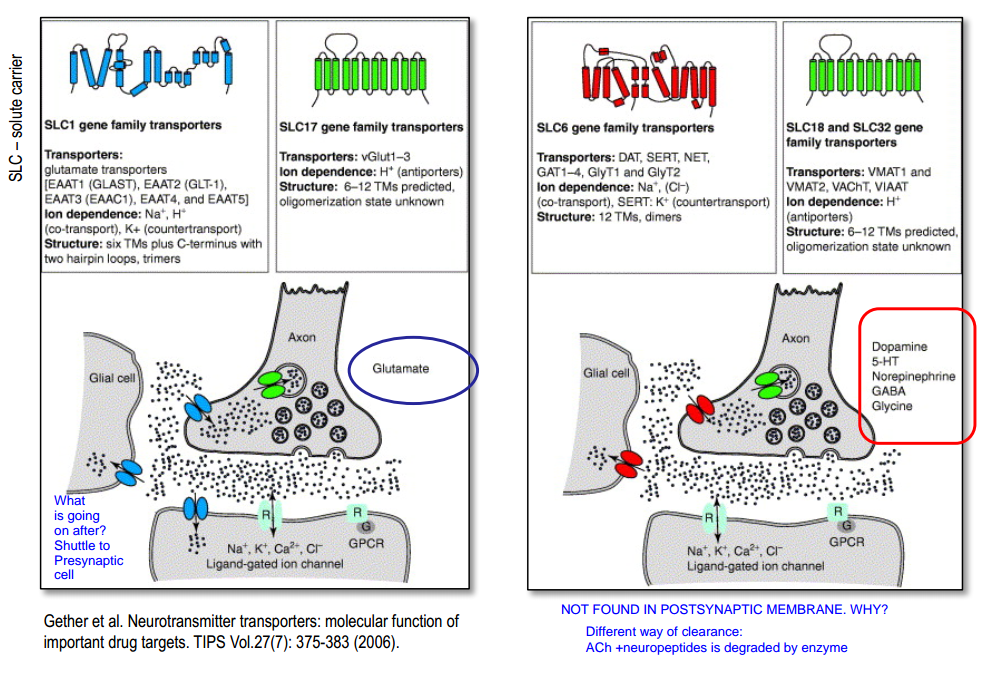
Glutamate-transporter
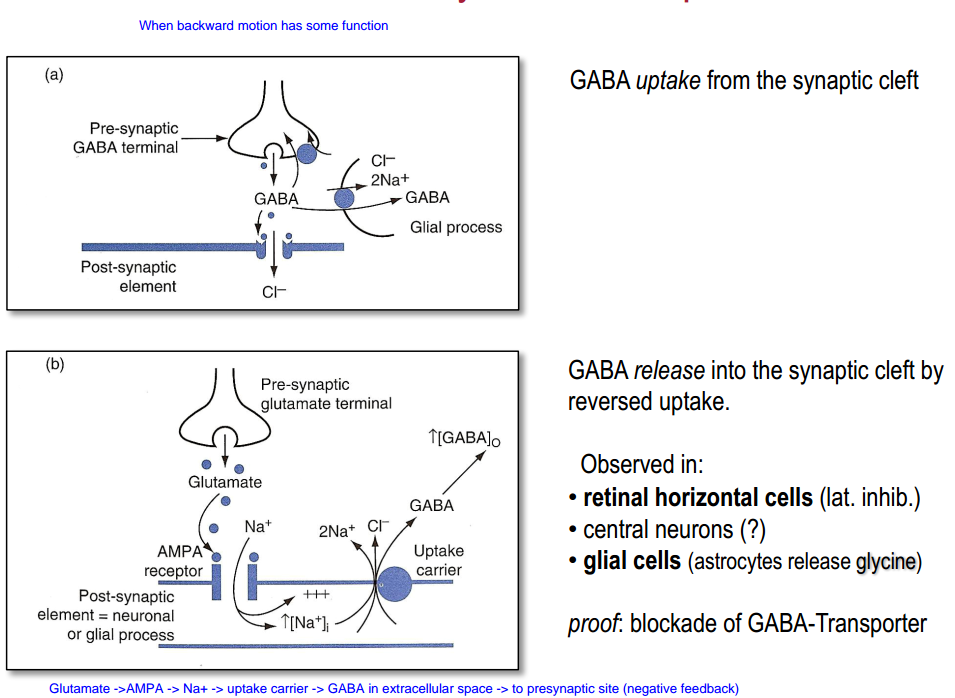
“Cooperative” binding:
- $3Na^+$, $H^+$, $Glu$ in $\rightarrow$ change in conformation $\rightarrow$ $K^+$ out $\rightarrow$ change in conformation $\rightarrow$ ...
Part 3. NT release by “reversed uptake”¶
- Transporter can always work in different direction if the concentrations changed dramatically (for example not enough energy for $Na^+-K^+$ pump)
Glutamate (pathological conditions)
GABA (negative feedback)
Part 4. Clinical relevance¶
- Dopamine-Transporter & Addiction
- Cocaine block Dopamine(DA) transporter $\rightarrow$ re-uptake of DA is blocked $\rightarrow$ increased concentration of DA $\rightarrow$ "reward" (in NA) $\rightarrow$ the reinforcing effects finally leading to cocaine addiction
- Serotonin-Transporter & Depression
- several psychiatric and affective / emotional disorders are caused by a malfunctioning serotonergic system / SERTs (depression, anxiety disorders, obsessive-compulsive disorders, …)
- treatment with drugs that block the uptake of 5-HT $\rightarrow$ increased concentration of 5-HT in the synaptic cleft (for example, Prozac)
- GABA-Transporter & Epilepsy
- GABA-Transporters (GATs) are targets of anticonvulsive substances
- epileptic seizures can be suppressed by increasing cortical inhibition
- application of drugs that prevent the uptake of GABA from the synaptic cleft $\rightarrow$ increased concentration of GABA in the synapse $\rightarrow$ increased inhibition $\rightarrow$ strong anti-convulsive effects(for example, Tiagabine )
Lecture 5. Directional Transport in Neurons¶
Part 1. Introduction¶
Why do neurons need “transport mechanisms” ?
- Neurons are highly polorized.(axons + dendrites)
- Proteins, organelles, (mRNAs) has to travel from soma (where they are produced) to their specific locations
- Diffusion is slow $\rightarrow$ we need active transport
Axonal arborization - axons also has many branches, and can terminate in different places.
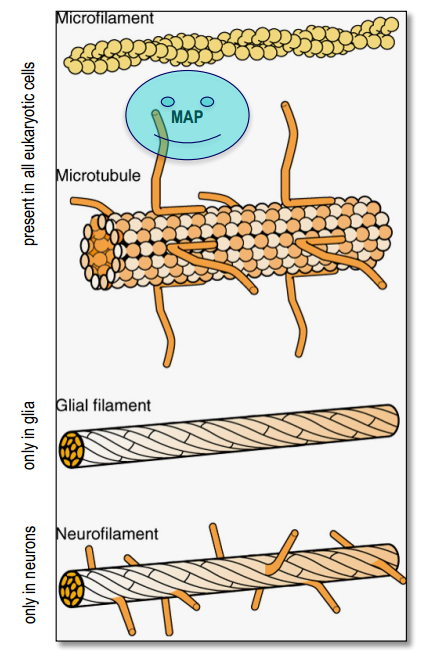
Part 2. Cytoskeletal elements¶
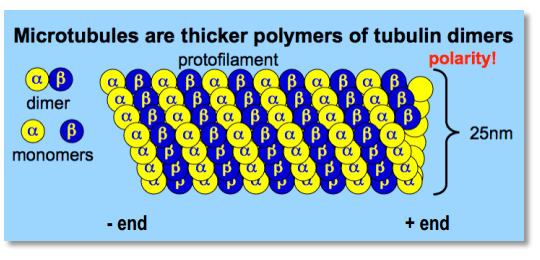
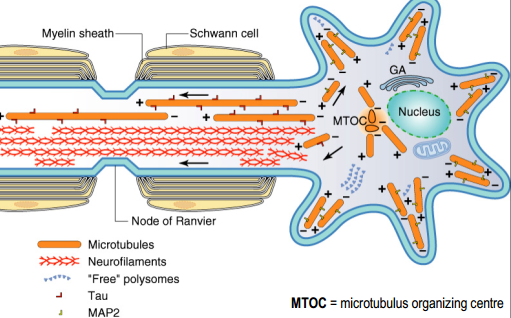
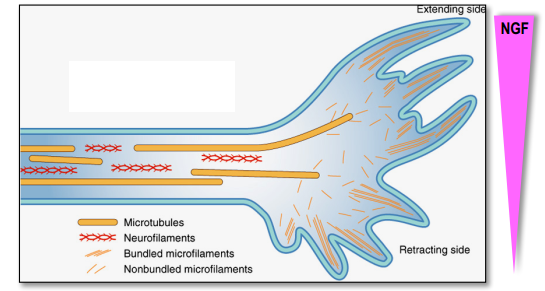
Microtubules
- cytoskeletal elements
- tracks for molecular motors proteins
- tick($25 nm$)
- polar because of alternating pattern of $\alpha$- and $\beta$-tubulin(molecular motors can recognize this polarity)
- +end grows much more rapidly then -end
- Axonal and dendritic microtubules differ in their orientation (+/-) and in their associated MAPs(Tau and MAP2)
- the growing of new parts of protein depends on concentration of some substances (Nerve growth factors)
Part 3. Molecular motors¶
Functions
- they power small-scale intracellular movements ([examples???])
- generate the forces responsible for muscle contraction
- move chromosomes during mitosis(replicated chromosomes are separated into two new nuclei)
- move enzymes along a DNA strand during transcription
- in neurons: move organelles along microtubule tracks
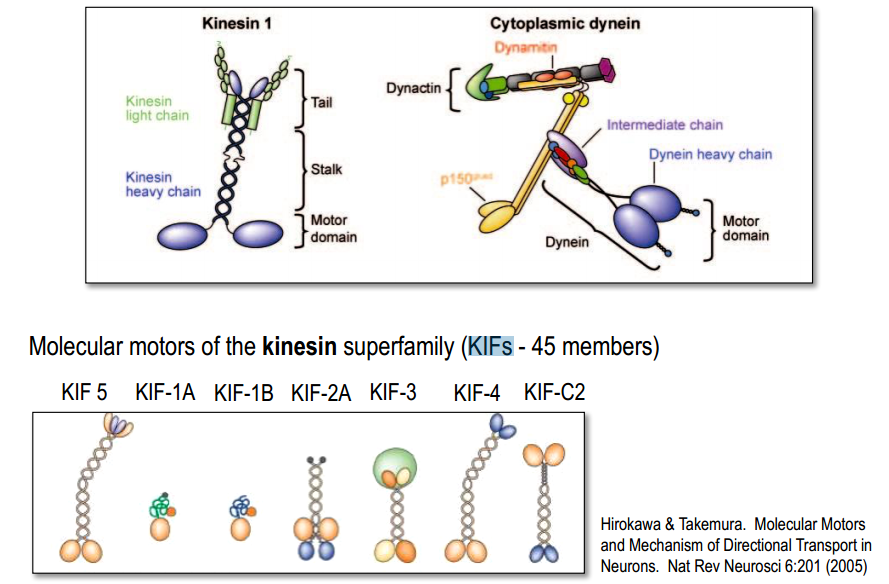
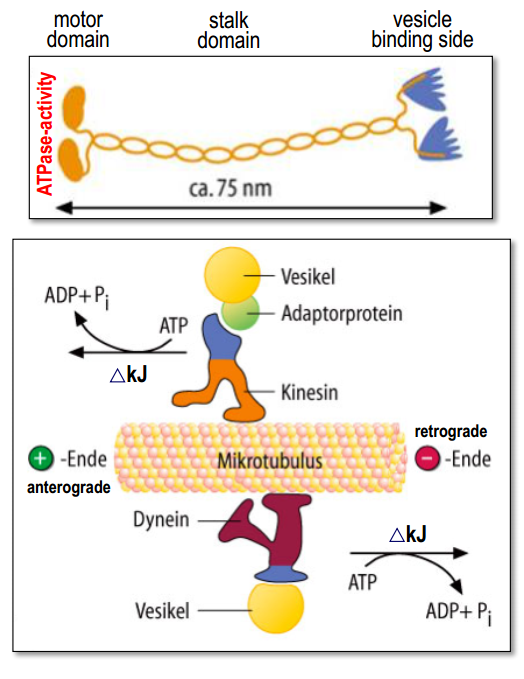
Molecular structure
insert_video('9RUHJhskW00',(0,4,50),(0,11,16))
- Upon contact with the microtubules, the motor domain hydrolysis ATP.
- The released energy induces a conformational change in the motor head which is then translated into a movement along the microtubule.
- kinesins moves cargo towards the +end (soma $\rightarrow$ axon terminal or dendrites)
- dyneins moves cargo towards the –end (axon terminal $\rightarrow$ soma).
- KIF‘s make use of adaptor protein complexes for cargo recognition and binding
- using different APC the same KIF can bing different substances
- direction of transport can be determine by different MAPs, and by different type of KIFs
What? Where? How quickly?
To axon terminal (by kinesins toward +end of microtubules)
- vesicles with NT-enzymes, peptide pre-coursers,
- vesicles with NT-receptors, ion channels, proteins for transmitter-release, docking molecules
- new lipids for membrane-turnover (20-40 cm/day)
- mitochondria (regenerated, metabolically active) (5-10 cm/day)
- cyto-soluble proteins: metabolic / glycolysis enzymes (slow 0.2-0.8 cm/day)
- cytoskeletal / structural proteins: neurofilaments, microtubules, microfilaments (slow 0.2-0.8 cm/day)
To soma (by dyneins toward -end of microtubules)
- removal of misfolded or aggregated proteins (!), membrane patches, cellular debris (20-30 cm/day)
- molecules for retrograde communication (NGF), toxins, viruses (HIV, herpes, …) (20-30 cm/day)
- mitochondria (degenerated, metabolically inactive) (5-10 cm/day)
To dendrites (by dyneins toward -end of microtubules)
- Molecules associated with postsynaptic densities
- Neurotransmitter receptors (NMDA-R, AMPA-R)
- Ion channels
- RNA granules containing specific mRNAs(> synaptic plasticity, LTP)
Part 4. Examples¶
Dendritic transport and synaptic plasticity
- KIF17 overexpression in transgeníc mice enhances working / episodic memory and spatial learning!
- KIF17 & NMDA-R genes are coregulated
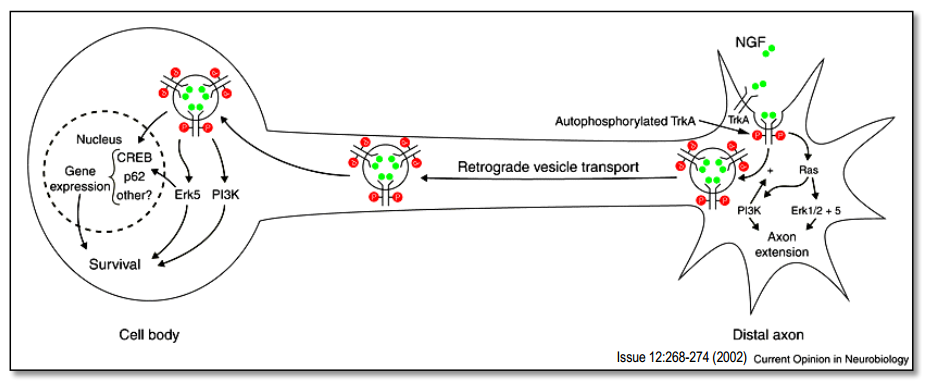
Retrograde communication of growth factors
- TyrosinKinase- (trk)-receptors bind to growth factors (e.g. NGF)
- growth and differentiation of neurons
- promote survival (= prevents apoptosis).
- trk-NGF-complexes are retrogradely transported to the soma and induce gene expression and, at the same time, suppresses apoptosis (‘programmed cell death’)
Retrograde transport of ‘neurotoxins'
Bacteria (Chlostridium tetani) enter an open wound:
- they produce and secrete tetanus toxin = protease
- taken-up (endocytosed) by axon terminals of motoneurons
- transported retrogradely to the soma (axonal transport !)
- reaches trans-synaptically glycinergic AT (spinal ventral horn)
- blocks glycine release (effect SNARE-complex) –> no inhibition any more
- tetanic muscle contractions
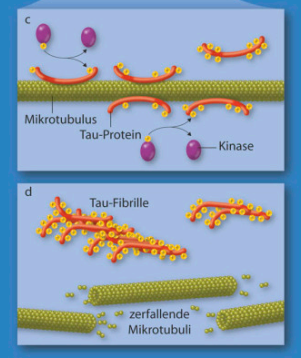
Axonal transport & neurodegenerative diseases
- Motor neuron disorders (ALS, motor neuropathy, muscular atrophy) (reduced ATPase activity $\rightarrow$ reduced motor activity)
- Alzheimer’s Disease (tangles = phosphorylated, aggregated tau-proteins (MAPs))
- Huntington’s Disease (motor-cargo-interaction)
- Parkinson’s Disease (mitochondrial damage $\rightarrow$ decreased ATP-supply in axons)
Lecture 6. Neurotransmitters¶
Part 1. Low molecular neurotransmitters vs Neuropeptides¶
Neurotransmitters & neuroactive substances
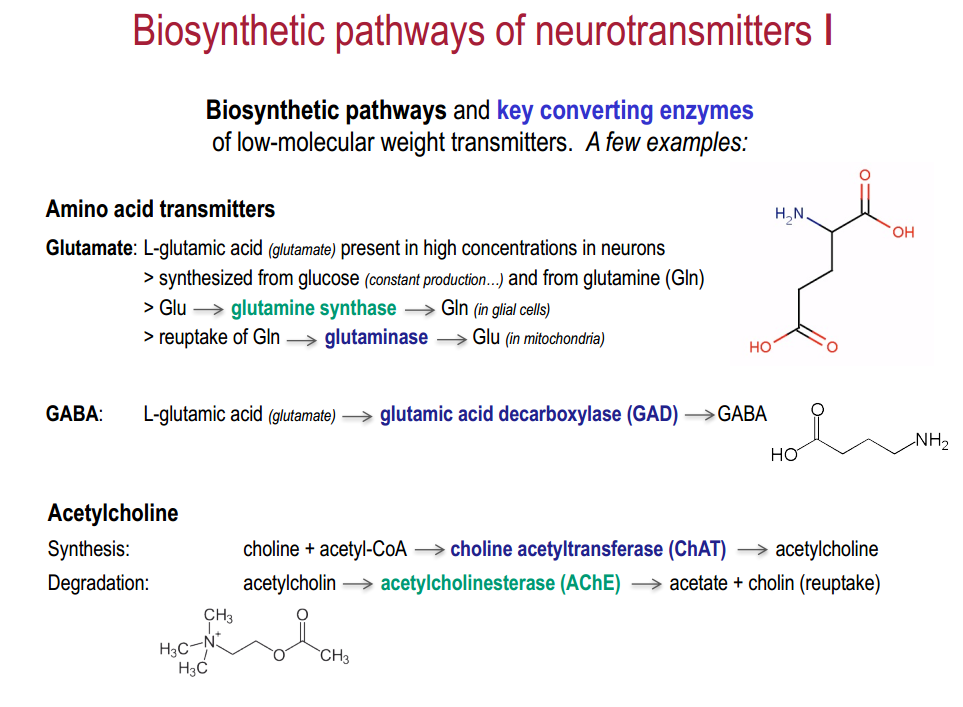
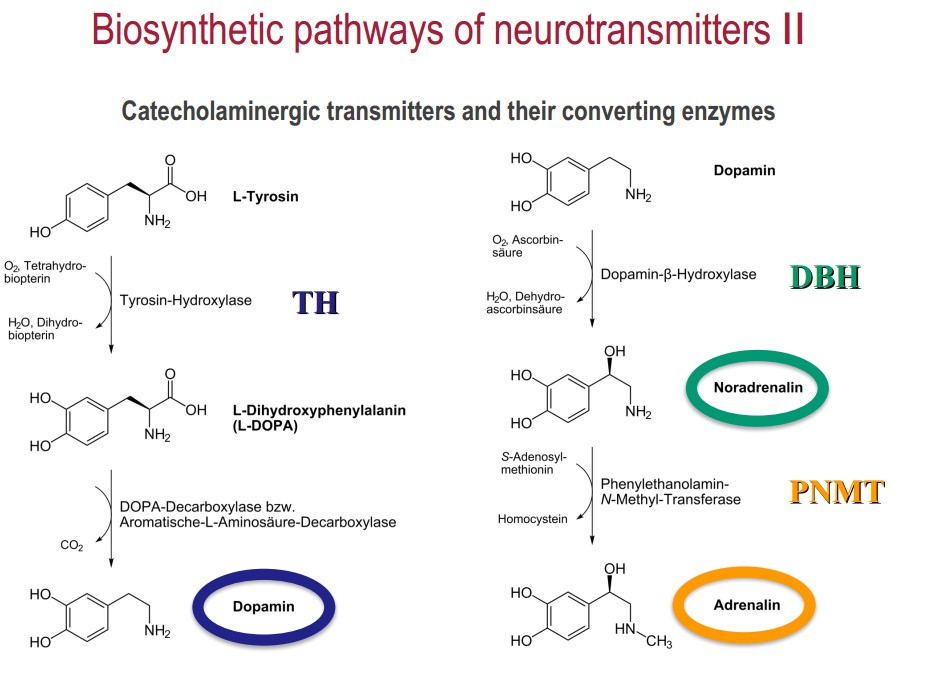
Low-molecular:
- amino acids: glutamate, GABA, glycine
- acetylcholin (motor endplate + central)
- biogenic amines: dopamine, nor-/adrenaline, serotonin, histamine
- others: ATP, adenosine, endocannabiniods
- glio-transmitter: D-serine
Neuropeptides (neuroactive peptides, >100):
- substance P, vasopressin, opioids, releasing-factors (pituitary….) sizes / amino acid residues: 5 (leu-enkephalin) – 39 (β-endorphin)
Gaseous neuroactive substances:
- nitric oxide / NO, carbon monoxide / CO
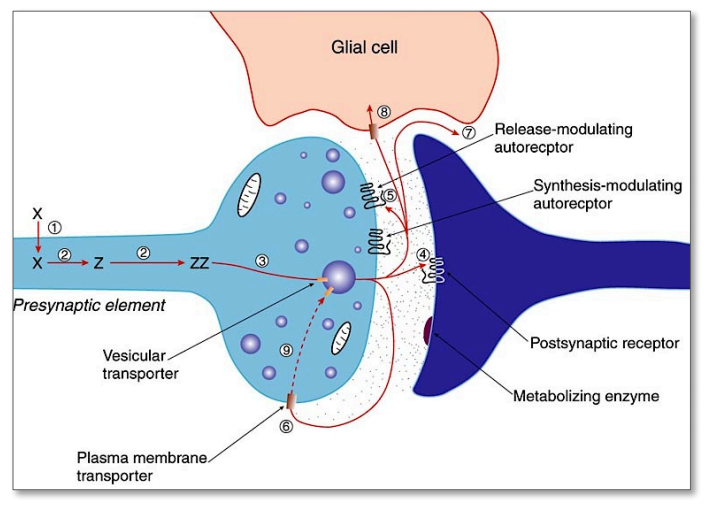
Low-molecular: Synthesis (1,2) $\rightarrow$ transport (3) $\rightarrow$ release (4,5)$\rightarrow$ (re)-uptake (6,7,8) $\rightarrow$ inactivation (9)
- Uptake and accumulation of precursor amino acids (X) into the neuron
- Precursor is sequentially metabolized (enzymes !!) to get the final transmitter molecule (Z)
- Transmitter is accumulated (ZZ) and pumped into the vesicles via vesicular transporters
4-5. Release into the synaptic cleft and binding to pre- and/or postsynaptic receptors
6-8. Diffusion or reuptake by transporters (exception: ACh is enzymatically degraded by AChE)
9 Metabolic inactivation / re-synthesis
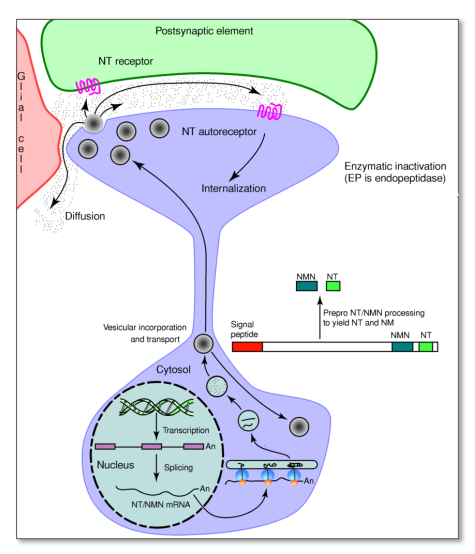
Peptides: synthesis $\rightarrow$ transport $\rightarrow$ release $\rightarrow$ termination
- Pre-cursor peptides are synthesized in the soma $\rightarrow$ packed in the Golgi-apparatus into secretory granules together with peptidases $\rightarrow$ granules transported to the terminal $\rightarrow$ the peptide pre-cursor is cleaved $\rightarrow$ active peptides in vesicles in axon terminal
- released from dense core vesicles in a Ca++-dependent fashion
- dense core vesicles are typically released at high firing-frequencies or burst-firing pattern
- neuropeptides are enzymatically inactivated by (rather unspecific) endopeptidases
- few released peptides are cleaved into more ‘active forms’ (angiotensin I angiotensin II and III)
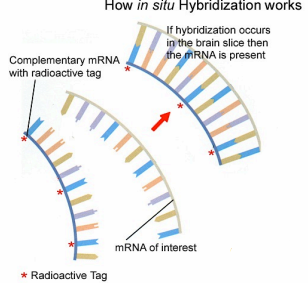
Methods of NT imaging
We look for some indicators of the NT:
- neurotransmitter itself (difficulty: size of molecules, not a problem for peptides)
- key converting enzymes (TH, DBH, GAD, ChAT) [Someone who will be NT in the future]
- vesicular NT-transporter (vGluT1-3, vGaT, …) [Someone who add NT to vesicles ]
Then we look for presence of mRNA of these indicators using in situ hybridization:
- mRNAs coding for key converting enzymes
- mRNAs coding for neuropeptides
- mRNAs coding for vesicular transporters
Pathways of synthesis
These NT are connected, so we get one from other
Part 2. Co-localization of Classical Transmitters & Neuropeptides¶
Dale’s principle of neurotransmitter release
Neurons release the same neurotransmitters from all of their axon terminals.
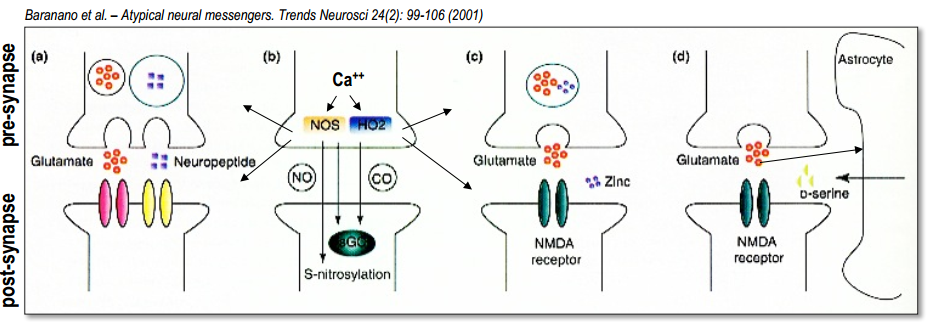
Part 3. Retrograde signaling¶
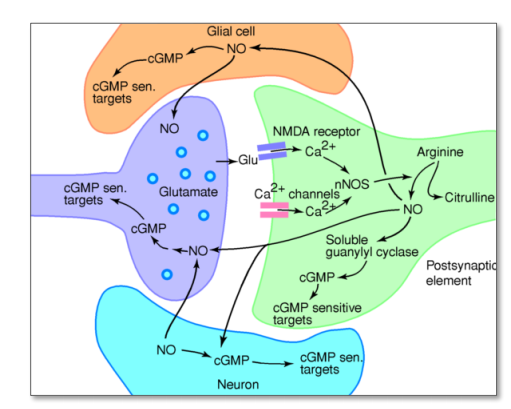
NO
- Glu $\rightarrow$ NMDA $\rightarrow$ $Ca^{2+}$ release $\rightarrow$ active enzyme nitric oxide-synthase ($nNOS$) $\rightarrow$ NO is generated from arginine $\rightarrow$ NO diffuses freely to Glia and axon terminal $\rightarrow$ NO activates an enzyme ($sGC$) $\rightarrow$ synthesizes the sec.mess. cGMP $\rightarrow$ cGMP-dependent target proteins (ion channels and protein kinase)
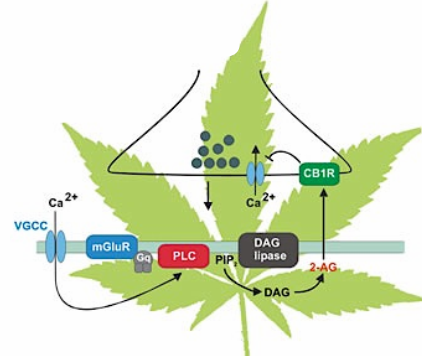
Endocannabinoids (2-AG, anandamide)
$Ca^{2+}$ or activated G-protein $\rightarrow$PLC (enzyme, cleave phospholipids )$\rightarrow$ PIP$_2$ $\rightarrow$ DAG $\rightarrow$ production of 2-AG(highly lipophilic and membrane permeable) $\rightarrow$ released into the extracellular space $\rightarrow$ binds specifically to $CB_1$-receptors located on presynaptic terminals $\rightarrow$ activation of $G_{i/o}$-protein $\rightarrow$ inhibition of $Ca^{2+}$ receptors $\rightarrow$ less NT (negative feedback)
Part 4. Neurotransmitter system¶
Principles
- often comprise a small set of well localized neurons (several thousands)
- many of them are found in lower brain areas / brain stem
- each neuron can influence many others through widespread axonal arbors
- modulate large assemblies of neurons (cortex, thalamus, spinal cord)
- axon terminals often release transmitter into extracellular space (hormone-like communication) (without cleft)
- these neurotransmitter systems often have general ‘regulatory’ functions (see cartoon …)
- change general physiological features, such as ‘excitability’, more or less ‘synchronously active’
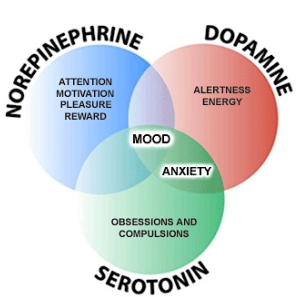
The noradrenergic system (LC, Brainstem noradrenergic cells)
LC
- projects to just about every part of the brain !
- involved in regulating arousal, attention, sleep-wake cycle, anxiety, mood, …
- LC is activated by ‘new, unexpected’ stimuli
- LC increases the brains ‘responsiveness’, ‘alertness’
Brainstem noradrenergic cells
- projects to autonomic cell groups, hypothalamus
- regulates autonomic / respiratory
The dopaminergic system (SN, VTA)
SN
- projects to basal ganglia
- involved in regulating motor functions
- facilitates the initiation of voluntary movements (Parkinson’s disease)
VTA
- projects to frontal cortex, limbic system
- crucial part of the brain’s reward system
- reinforcement of adaptive behaviors
The serotonergic system(Raphe nuclei)
Rostral raphe nuclei
- projects to various forebrain areas, particularly cortex
- with LC-projections, part of the ‘ascending reticular activating system’
- arousal of the brain, sleep-wake cycle as well as different stages of sleep
- regulation of mood, emotional behaviors (affective disorders)
Caudal raphe nuclei
- projects to spinal cord
- modulation of pain-related sensory signals
The cholinergic system (BF)
Basal forebrain projections (BF):
- medial septal nuclei $\rightarrow$ hippocampus
- basal nucleus of Meynert $\rightarrow$ cortex
- cognitive functions – learning & memory
- cells degenerate in Alzheimer’s patients
- arousal, sleep-wake-cycle
Signal Transduction - General¶
Ionotropic vs. metabotropic receptors:
- General molecular composition of these receptor types.
- Difference between cation vs. anion channels (ACh-/GABA$_A$-rec.).
- General features of ionotropic vs. metabotropic receptors (time course, effects, duration of effects, amplification, cross-talk).
- Metabotropic receptor: signal transduction from extracellular to intracellular – sec. mess. cascades – ‘long’ vs. ‘short’ cascades.
$Ca^{2+}$ functions in excitable cells
- Name a few functions of $Ca^{2+}$ in neurons …
- Name the major structures/mechanisms by which $Ca^{2+}$ enters the cytoplasm.
- Mechanisms by which $Ca^{2+}$ action is ceased/buffered and mechanisms of $Ca^{2+}$ removal.
GABA receptors¶
GABA$_A$-receptors
- Subunits / subunit composition – function …
- Distinctive features of GABA$_A$-receptors – pharmacology.
- Physiological effect of GABA$_A$ activation.
- Potentiation of GABA$_A$-currents.
GABA$_B$-receptors
- Molecular composition / VFTM / hetero-dimer.
- Occurrence in the CNS.
- Second messenger coupling:
- Pre-synaptic localization – effect (in detail).
- Post-synaptic localization – effect (in detail)
Glutamate Receptors¶
Ionotropic glutamate-receptors
- Molecular composition – gating / compare.
- Characterize the two classes of iGlu-receptors.
- Major function of AMPA-receptors.
- Distinctive features of NMDA-receptors (in detail).
- Gating mechanisms / major function.
Metabotropic glutamate-receptors
- 8 types – 3 groups / how are the “Groups” characterized?
- Second messenger coupling Group II and III.
- Second messenger coupling Group I (in detail).
- Occurrence of mGlu-Rs in the CNS.
Neurotransmitter Transporters¶
- Why do neurons need ‘trans-membrane transport mechanisms’ ?
- Describe the working principles of ‘pumps’ vs. ‘transporters’.
- Where in the neuropil are NT-transporters located?
- Why are NT-transporters crucial for chemical neurotransmission?
- Example of NT-transporters acting as ‘drug’ targets.
Directional Transport¶
Mechanisms of directional transport in neurons
- Why do neurons need active, directional transport mechanisms?
- Which of the cytoskeletal elements serves as tracks / ‘polarity’?
- What is a ‘molecular motor’ and how does it work?
- Which ‘types’ of molecular motors are crucial for neuronal transport?
- Give examples of fast anterogradely transported cargoes.
- Give examples of fast retrogradely transported cargoes.
- What are typical cargoes of the dendritic transport?
- How do molecular motors recognize / bind their specific cargo?
Neurotransmitters (NTs), peptides and neuroactive substances¶
• Give a few examples of classical NTs, neuropeptides and other neuroactive substances. • Do NTs/peptides always act on both, ionotropic and metabotropic receptors? Give an example … • What is meant with ‘co-localization’ of transmitters – which forms of co-localization do you know? • Describe one ‘retrograde’ signaling system (NO-/nitric oxid-or endocannabinoid-system). • Which methods can be applied to demonstrate NTs/peptides in neuronal tissue? What is the major difference between the two methods discussed in lecture? • What is meant with ‘diffuse modulatory neurotransmitter systems’ ? Give one example (NT – localization – function)
Questions
Lecture 1. What is the function of rapsyn?
Lecture 8. Glia¶